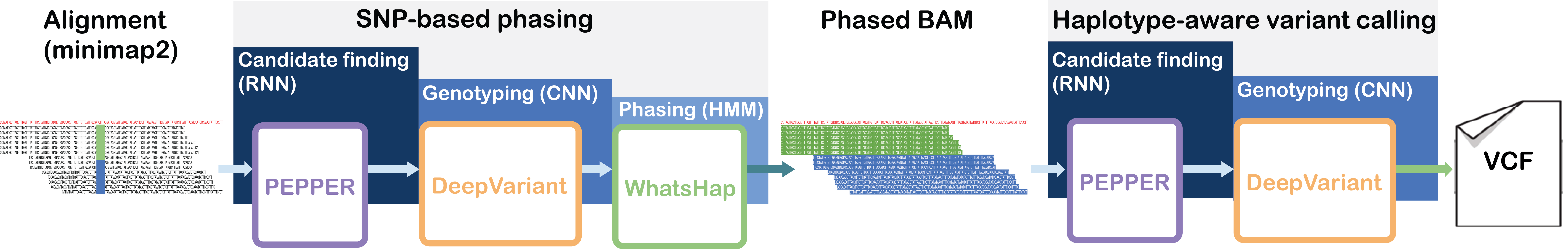
In collaboration with the DeepVariant group, we are developing a haplotype-aware variant calling pipeline for Oxford Nanopore sequencing technology. We are releasing the pipeline with limited support until we finalize the pipeline.
- PrecisionFDA truth challenge v2 results
- Limited support statement
- Requirements
- ONT HG002 chr20 case-study
- ONT Whole-genome evaluations
- How to run
- Quickstart
- Authors
We participated in the PrecisionFDA truth challenge V2 and won two awards in the ONT category.
A public table with more detailed performance metrics for all submissions is available here and a details of the table values can be found here.
Here we summarize the variant calling performance of the winning submissions of Illumina and ONT in all benchmark regions and Difficult-to-map regions category. The numbers are directly lifted from the pFDA results table.
| Type | Team | Technology | Pipeline | True positives |
False negatives |
False positives |
Recall | Precision | F1-Score |
|---|---|---|---|---|---|---|---|---|---|
| SNP | UCSC CGL-Google Health | ONT | PEPPER-DeepVariant | 257460 | 3841 | 2465 | 0.98528 | 0.99045 | 0.98785 |
| SNP | DRAGEN | Illumina | DRAGEN | 251058 | 10245 | 5610 | 0.96078 | 0.97811 | 0.96936 |
| INDEL | UCSC CGL-Google Health | ONT | PEPPER-DeepVariant | 13815 | 2673 | 670 | 0.83780 | 0.95408 | 0.89217 |
| INDEL | DRAGEN | Illumina | DRAGEN | 15883 | 605 | 420 | 0.96322 | 0.97447 | 0.96881 |
| Type | Team | Technology | Pipeline | True positives |
False negatives |
False positives |
Recall | Precision | F1-Score |
|---|---|---|---|---|---|---|---|---|---|
| SNP | UCSC CGL-Google Health | ONT | PEPPER-DeepVariant | 3328756 | 14716 | 8914 | 0.99558 | 0.99731 | 0.99645 |
| SNP | DRAGEN | Illumina | DRAGEN | 3331542 | 11973 | 7575 | 0.99641 | 0.99773 | 0.99707 |
| INDEL | UCSC CGL-Google Health | ONT | PEPPER-DeepVariant | 320171 | 187923 | 52012 | 0.63009 | 0.86333 | 0.72849 |
| INDEL | DRAGEN | Illumina | DRAGEN | 506539 | 1588 | 1437 | 0.99687 | 0.99728 | 0.99708 |
We are releasing this pipeline with limited support as we are actively developing the pipeline. This pipeline is developed outside the main repository of DeepVariant. If you have issues with running this pipeline, please open a github issue in this repository (PEPPER). We will update this page as we evaluate the pipeline across all platforms.
We are releasing this pipeline executable only via docker. Other executables will be provided as we finalize the pipeline.
We expect the input files meet the following requirements to produce results that we have reported.
Alignment file (BAM):
Coverage: ~50-80x
Pore: R9.4.1
Basecaller: Guppy 3.6.0
Platform: PromethION or MinION
Reference file:
Standard release of GRCh38 or GRCh37.
We have not tested this pipeline on references with smaller contigs i.e. assemblies.
We have not yet tested the pipeline on different pores and devices. We will update this document as we validate the pipeline.
As we use WhatsHap to phase the BAM file, please expect disk-space usage of ~4x the size of the BAM file.
NOTE: This is a chromosome-level case-study. Please see the Quickstart section of this document for demonstration of this pipeline on a smaller dataset.
We evaluated this pipeline on ~50x HG002 data. The data is publicly available, please feel free to download, run and evaluate the pipeline.
Sample: HG002
Coverage: ~50-80x
Basecaller: Guppy 3.6.0
Region: chr20
Reference: GRCh38_no_altPlease install docker and wget if you don't have it installed already.
# Install wget to download data files.
sudo apt-get -qq -y update
sudo apt-get -qq -y install wget
# Install docker using instructions on:
# https://docs.docker.com/install/linux/docker-ce/ubuntu/
sudo apt-get -qq -y install apt-transport-https ca-certificates curl gnupg-agent software-properties-common
curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo apt-key add -
sudo add-apt-repository \
"deb [arch=amd64] https://download.docker.com/linux/ubuntu \
$(lsb_release -cs) \
stable"
sudo apt-get -qq -y update
sudo apt-get -qq -y install docker-ce
docker --versionBASE="${HOME}/ont-case-study"
# Set up input data
INPUT_DIR="${BASE}/input/data"
REF="GRCh38_no_alt_chr20.fa"
BAM="HG002_prom_R941_guppy360_2_GRCh38_ch20.bam"
TRUTH_VCF="HG002_GRCh38_GIABv4.1.vcf.gz"
TRUTH_BED="HG002_GRCh38_1_22_v4.1_draft_benchmark.bed"
# Set the number of CPUs to use
THREADS="64"
# Set up output directory
OUTPUT_DIR="${BASE}/output"
OUTPUT_VCF="PEPPER_HP_DEEPVARIANT_FINAL_OUTPUT.vcf.gz"
## Create local directory structure
mkdir -p "${OUTPUT_DIR}"
mkdir -p "${INPUT_DIR}"
# Download the data to input directory
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-casestudy/HG002_prom_R941_guppy360_2_GRCh38_ch20.bam
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-casestudy/HG002_prom_R941_guppy360_2_GRCh38_ch20.bam.bai
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-casestudy/GRCh38_no_alt_chr20.fa
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-casestudy/GRCh38_no_alt_chr20.fa.fai
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-casestudy/HG002_GRCh38_GIABv4.1.vcf.gz
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-casestudy/HG002_GRCh38_1_22_v4.1_draft_benchmark.bed
## Pull the docker image.
sudo docker pull kishwars/pepper_deepvariant_cpu:latest
# Run PEPPER-DeepVariant High-Accuracy mode on CPU
# For other versions and GPU specific version, please consult the How to run section of this document
sudo docker run --ipc=host \
-v "${INPUT_DIR}":"${INPUT_DIR}" \
-v "${OUTPUT_DIR}":"${OUTPUT_DIR}" \
kishwars/pepper_deepvariant_cpu:latest \
/opt/run_pepper_deepvariant.sh \
-f "${INPUT_DIR}/${REF}" \
-b "${INPUT_DIR}/${BAM}" \
-o "${OUTPUT_DIR}" \
-t ${THREADS} \
-x 1You can evaluate the variants using hap.py.
# Pull the docker image
sudo docker pull pkrusche/hap.py:latest
# Run hap.py
sudo docker run -it \
-v "${INPUT_DIR}":"${INPUT_DIR}" \
-v "${OUTPUT_DIR}":"${OUTPUT_DIR}" \
pkrusche/hap.py:latest \
/opt/hap.py/bin/hap.py \
${INPUT_DIR}/${TRUTH_VCF} \
${OUTPUT_DIR}/${OUTPUT_VCF} \
-f "${INPUT_DIR}/${TRUTH_BED}" \
-r "${INPUT_DIR}/${REF}" \
-o "${OUTPUT_DIR}/happy.output" \
--pass-only \
-l chr20 \
--engine=vcfeval \
--threads="${THREADS}"You can run this pipeline on run-time efficient mode or high-accuracy mode (high-accuracy mode enabled by setting parameter -x 1). The difference between these two modes is the haplotype-aware model used for DeepVariant. The high-accuracy mode uses a better feature representation known as rows in DeepVariant that results in higher accuracy but slower run-time.
We have tested the high-accuracy and run-time efficient mode of this pipeline on ~50x HG002 chr20 data.
Compute node description:
CPUs:
CPU(s): 32
CPU MHz: 1979.549
CPU max MHz: 3400.0000
CPU min MHz: 2200.0000
GPU:
GPU(s): 1
GPU Model: GeForce GTX 1080ti
GPU Memory: 11177MiBRun-time analysis:
| Mode | Platform | Sample | Region | Coverage | Run-time (wall-clock) |
|---|---|---|---|---|---|
| Run-time | CPU | HG002 | chr20 | ~50x | 118.34 mins |
| Run-time | GPU | HG002 | chr20 | ~50x | 71.03 mins |
| High-accuracy | CPU | HG002 | chr20 | ~50x | 211.92 mins |
| High-accuracy | GPU | HG002 | chr20 | ~50x | 164.71 mins |
Variant-calling performance:
| Mode | Type | Truth Total | True positives | False negatives | False positives | Recall | Precision | F1-Score |
|---|---|---|---|---|---|---|---|---|
| Run-time | INDEL | 11271 | 6318 | 4953 | 1826 | 0.560554 | 0.778935 | 0.651942 |
| Run-time | SNP | 71334 | 71037 | 297 | 191 | 0.995836 | 0.997319 | 0.996577 |
| High-accuracy | INDEL | 11271 | 6393 | 4878 | 1556 | 0.567208 | 0.80733 | 0.666295 |
| High-accuracy | SNP | 71334 | 71054 | 280 | 209 | 0.996075 | 0.997068 | 0.996571 |
We evaluated our pipeline on three samples (HG002-HG003-HG004). At the time of this release, only v3.3.2 truth was available for HG003 and HG004. Data description:
Sample: HG002, HG003, HG004
Coverage: ~50x, ~80x, ~80x
Basecaller: Guppy 3.6.0
Region: WGS
Referrence: GRCh38_no_altHere we report the whole genome performance on three samples.
| Reference | Truth | Sample | Coverage | Type | Recall | Precision | F1 Score |
|---|---|---|---|---|---|---|---|
| GRCh37 | GIAB v3.3.2 | HG002 | ~45-55x | SNV | 0.99605 | 0.996219 | 0.996135 |
| GRCh37 | GIAB v3.3.2 | HG003 | ~75-85x | SNV | 0.99546 | 0.997782 | 0.99662 |
| GRCh37 | GIAB v3.3.2 | HG004 | ~75-85x | SNV | 0.996939 | 0.996764 | 0.996852 |
We have combined all of the steps to run sequentially using one script. You can run the pipeline on your CPU-only or GPU machines using the command-line instructions provided here. Please see Quickstart section or HG002 chr20 case-study which provides example datasets.
Running the variant calling pipeline is as simple as: On a CPU-based machine:
# For Run-time efficient mode:
docker run --ipc=host \
-v /input_dir:/input_dir \
-v /output_dir:/output_dir \
kishwars/pepper_deepvariant_cpu:latest \
/opt/run_pepper_deepvariant.sh \
-b </input/bam_file.bam> \
-f </input/reference_file.fasta> \
-o </output/output_dir/> \
-t <number_of_threads>
# For High-Accuracy mode:
docker run --ipc=host \
-v /input_dir:/input_dir \
-v /output_dir:/output_dir \
kishwars/pepper_deepvariant_cpu:latest \
/opt/run_pepper_deepvariant.sh \
-b </input/bam_file.bam> \
-f </input/reference_file.fasta> \
-o </output/output_dir/> \
-t <number_of_threads> \
-x 1On a GPU based machine (make sure to install nvidia-docker):
# For Run-time efficient mode:
docker run --ipc=host \
-v /input_dir:/input_dir \
-v /output_dir:/output_dir \
--gpus all \
kishwars/pepper_deepvariant_gpu:latest \
/opt/run_pepper_deepvariant.sh \
-b </input/bam_file.bam> \
-f </input/reference_file.fasta> \
-o </output/output_dir/> \
-t <number_of_threads>
# For High-Accuracy mode:
docker run --ipc=host \
-v /input_dir:/input_dir \
-v /output_dir:/output_dir \
--gpus all \
kishwars/pepper_deepvariant_gpu:latest \
/opt/run_pepper_deepvariant.sh \
-b </input/bam_file.bam> \
-f </input/reference_file.fasta> \
-o </output/output_dir/> \
-t <number_of_threads> \
-x 1Here is an example on how to run the pipeline on a small example. Please see the HG002 chr20 case-study section of this document for a chromosome level evaluation.
Please install docker and wget if you don't have it installed already.
# Install wget to download data files.
sudo apt-get -qq -y update
sudo apt-get -qq -y install wget
# Install docker using instructions on:
# https://docs.docker.com/install/linux/docker-ce/ubuntu/
sudo apt-get -qq -y install apt-transport-https ca-certificates curl gnupg-agent software-properties-common
curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo apt-key add -
sudo add-apt-repository \
"deb [arch=amd64] https://download.docker.com/linux/ubuntu \
$(lsb_release -cs) \
stable"
sudo apt-get -qq -y update
sudo apt-get -qq -y install docker-ce
docker --versionBASE="${HOME}/ont-quickstart"
# Set up input data
INPUT_DIR="${BASE}/input/data"
REF="GRCh38_no_alt_chr20.fa"
BAM="HG002_prom_R941_guppy360_2_GRCh38_TEST.bam"
TRUTH_VCF="HG002_GRCh38_GIABv4.1.TRUTH.TEST.vcf.gz"
TRUTH_BED="HG002_GRCh38_GIABv4.1.TRUTH.TEST.bed"
# Set the number of CPUs to use
THREADS="1"
# Set up output directory
OUTPUT_DIR="${BASE}/output"
OUTPUT_VCF="PEPPER_HP_DEEPVARIANT_FINAL_OUTPUT.vcf.gz"
## Create local directory structure
mkdir -p "${OUTPUT_DIR}"
mkdir -p "${INPUT_DIR}"
# Download the data to input directory
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-quickstart/HG002_prom_R941_guppy360_2_GRCh38_TEST.bam
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-quickstart/HG002_prom_R941_guppy360_2_GRCh38_TEST.bam.bai
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-quickstart/GRCh38_no_alt_chr20.fa
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-quickstart/GRCh38_no_alt_chr20.fa.fai
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-quickstart/HG002_GRCh38_GIABv4.1.TRUTH.TEST.bed
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-quickstart/HG002_GRCh38_GIABv4.1.TRUTH.TEST.vcf.gz
wget -P ${INPUT_DIR} https://storage.googleapis.com/kishwar-helen/ont-quickstart/HG002_GRCh38_GIABv4.1.TRUTH.TEST.vcf.gz.tbi
## Pull the docker image.
sudo docker pull kishwars/pepper_deepvariant_cpu:latest
# Run PEPPER-DeepVariant High-Accuracy mode on CPU
sudo docker run --ipc=host \
-v "${INPUT_DIR}":"${INPUT_DIR}" \
-v "${OUTPUT_DIR}":"${OUTPUT_DIR}" \
kishwars/pepper_deepvariant_cpu:latest \
/opt/run_pepper_deepvariant.sh \
-f "${INPUT_DIR}/${REF}" \
-b "${INPUT_DIR}/${BAM}" \
-o "${OUTPUT_DIR}" \
-t ${THREADS} \
-r chr20:1000000-1020000 \
-x 1
# For run-time efficient mode on CPU-only machines:
sudo docker run --ipc=host \
-v "${INPUT_DIR}":"${INPUT_DIR}" \
-v "${OUTPUT_DIR}":"${OUTPUT_DIR}" \
kishwars/pepper_deepvariant_cpu:latest \
/opt/run_pepper_deepvariant.sh \
-f "${INPUT_DIR}/${REF}" \
-b "${INPUT_DIR}/${BAM}" \
-o "${OUTPUT_DIR}" \
-t ${THREADS} \
-r chr20:1000000-1020000
# On GPU machines with high-accuracy (please install nvidia-docker to use this)
## Pull the docker image.
sudo docker pull kishwars/pepper_deepvariant_cpu:latest
# Run PEPPER-DeepVariant High-Accuracy mode on GPU
sudo docker run --ipc=host \
--gpus all \
-v "${INPUT_DIR}":"${INPUT_DIR}" \
-v "${OUTPUT_DIR}":"${OUTPUT_DIR}" \
kishwars/pepper_deepvariant_gpu:latest \
/opt/run_pepper_deepvariant.sh \
-f "${INPUT_DIR}/${REF}" \
-b "${INPUT_DIR}/${BAM}" \
-o "${OUTPUT_DIR}" \
-t ${THREADS} \
-x 1 \
-r chr20:1000000-1020000You can use hap.py to evaluate the variants.
# Pull the docker image
sudo docker pull pkrusche/hap.py:latest
# Run hap.py
sudo docker run -it \
-v "${INPUT_DIR}":"${INPUT_DIR}" \
-v "${OUTPUT_DIR}":"${OUTPUT_DIR}" \
pkrusche/hap.py:latest \
/opt/hap.py/bin/hap.py \
${INPUT_DIR}/${TRUTH_VCF} \
${OUTPUT_DIR}/${OUTPUT_VCF} \
-f "${INPUT_DIR}/${TRUTH_BED}" \
-r "${INPUT_DIR}/${REF}" \
-o "${OUTPUT_DIR}/happy.output" \
--pass-only \
-l chr20 \
--engine=vcfeval \
--threads="${THREADS}"Expected output:
| Type | Filter | TRUTH.TOTAL | TRUTH.TP | METRIC.Recall | METRIC.Precision | METRIC.F1_Score |
|---|---|---|---|---|---|---|
| INDEL | ALL | 3 | 2 | 0.666667 | 1.0 | 0.8 |
| SNP | ALL | 45 | 45 | 1.0 | 1.0 | 1.0 |
This pipeline is a collaboration between UCSC genomics institute and the genomics team at Google health.
- Kishwar Shafin
- Trevor Pesout
- Miten Jain
- Benedict Paten
- Pi-Chuan Chang
- Alexey Kolesnikov
- Maria Nattestad
- Gunjan Baid
- Sidharth Goel
- Howard Yang
- Andrew Carroll