Assisting Pathologists in Detecting Cancer with Deep Learning
March 3, 2017
Posted by Martin Stumpe, Technical Lead, and Lily Peng, Product Manager
A pathologist’s report after reviewing a patient’s biological tissue samples is often the gold standard in the diagnosis of many diseases. For cancer in particular, a pathologist’s diagnosis has a profound impact on a patient’s therapy. The reviewing of pathology slides is a very complex task, requiring years of training to gain the expertise and experience to do well.
Even with this extensive training, there can be substantial variability in the diagnoses given by different pathologists for the same patient, which can lead to misdiagnoses. For example, agreement in diagnosis for some forms of breast cancer can be as low as 48%, and similarly low for prostate cancer. The lack of agreement is not surprising given the massive amount of information that must be reviewed in order to make an accurate diagnosis. Pathologists are responsible for reviewing all the biological tissues visible on a slide. However, there can be many slides per patient, each of which is 10+ gigapixels when digitized at 40X magnification. Imagine having to go through a thousand 10 megapixel (MP) photos, and having to be responsible for every pixel. Needless to say, this is a lot of data to cover, and often time is limited.
To address these issues of limited time and diagnostic variability, we are investigating how deep learning can be applied to digital pathology, by creating an automated detection algorithm that can naturally complement pathologists’ workflow. We used images (graciously provided by the Radboud University Medical Center) which have also been used for the 2016 ISBI Camelyon Challenge1 to train algorithms that were optimized for localization of breast cancer that has spread (metastasized) to lymph nodes adjacent to the breast.
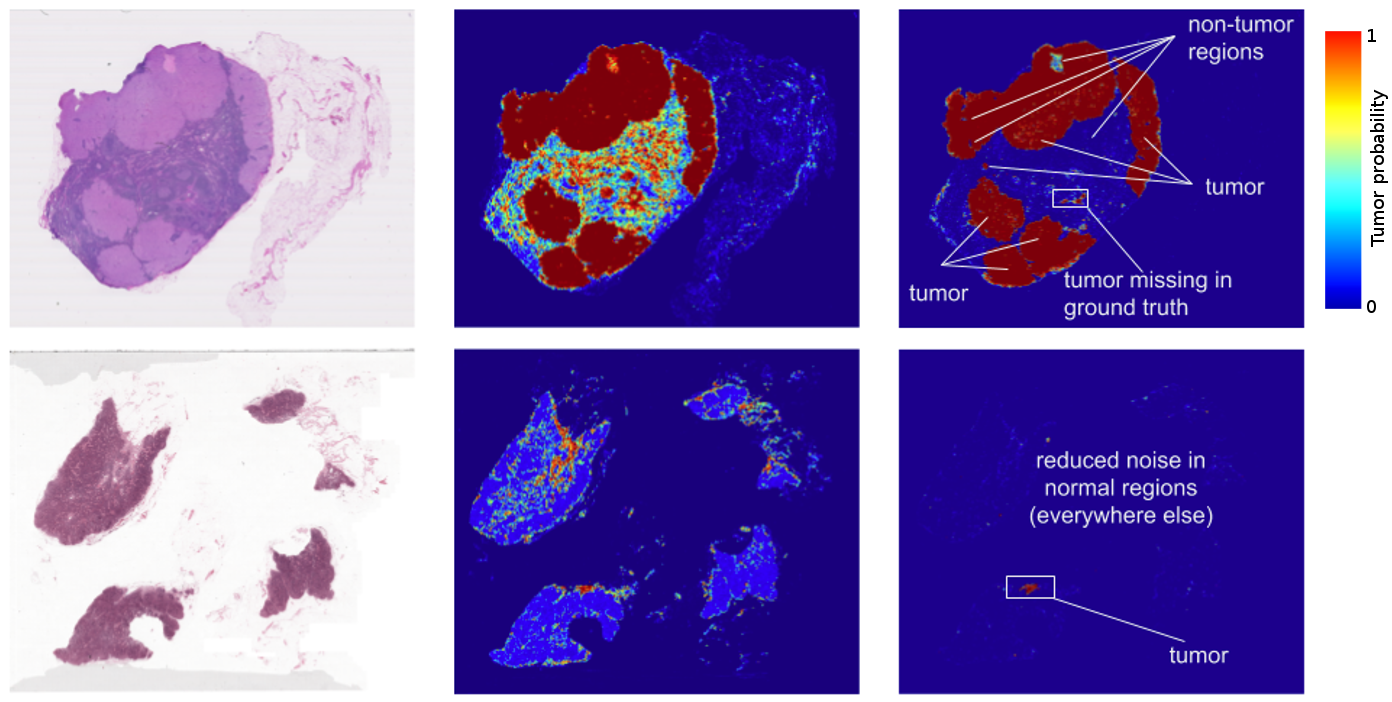
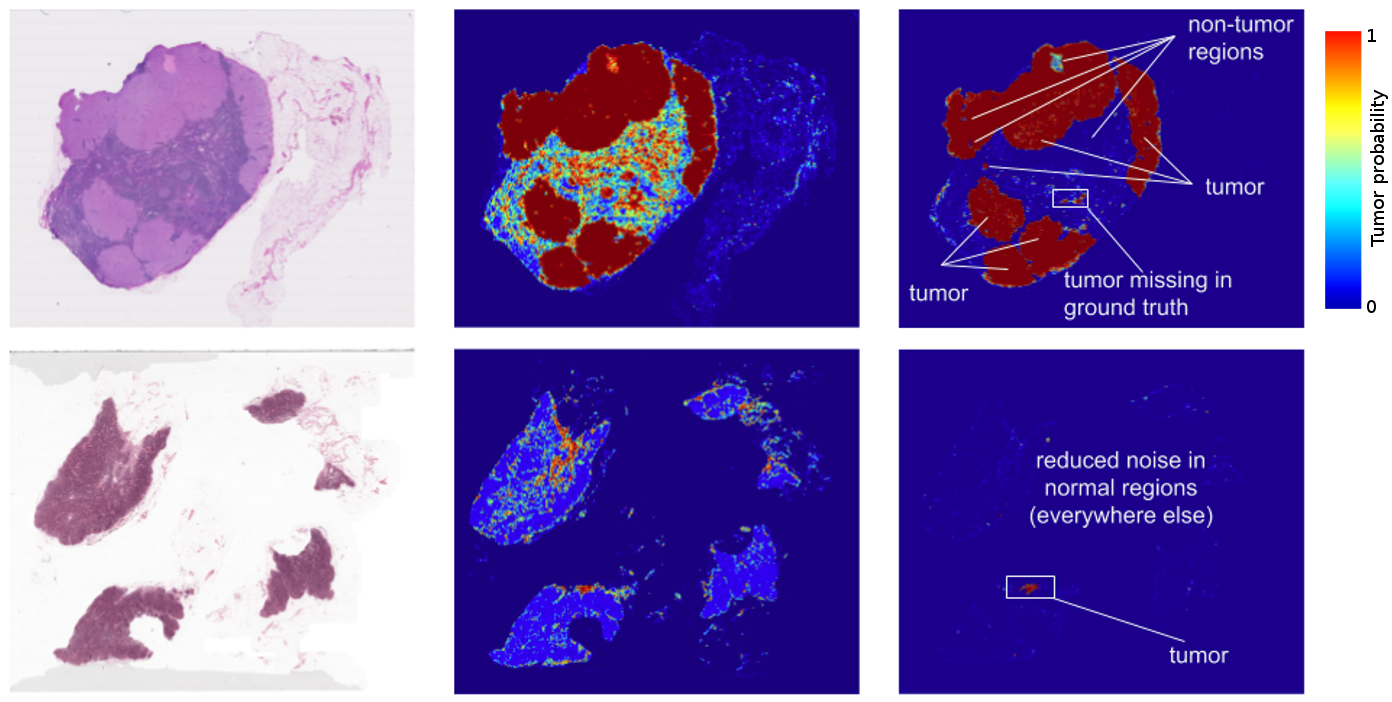
The results? Standard “off-the-shelf” deep learning approaches like Inception (aka GoogLeNet) worked reasonably well for both tasks, although the tumor probability prediction heatmaps produced were a bit noisy. After additional customization, including training networks to examine the image at different magnifications (much like what a pathologist does), we showed that it was possible to train a model that either matched or exceeded the performance of a pathologist who had unlimited time to examine the slides.
FROC) for the algorithm reached 89%, which significantly exceeded the score of 73% for a pathologist with no time constraint2. We were not the only ones to see promising results, as other groups were getting scores as high as 81% with the same dataset. Even more exciting for us was that our model generalized very well, even to images that were acquired from a different hospital using different scanners. For full details, see our paper “Detecting Cancer Metastases on Gigapixel Pathology Images”.
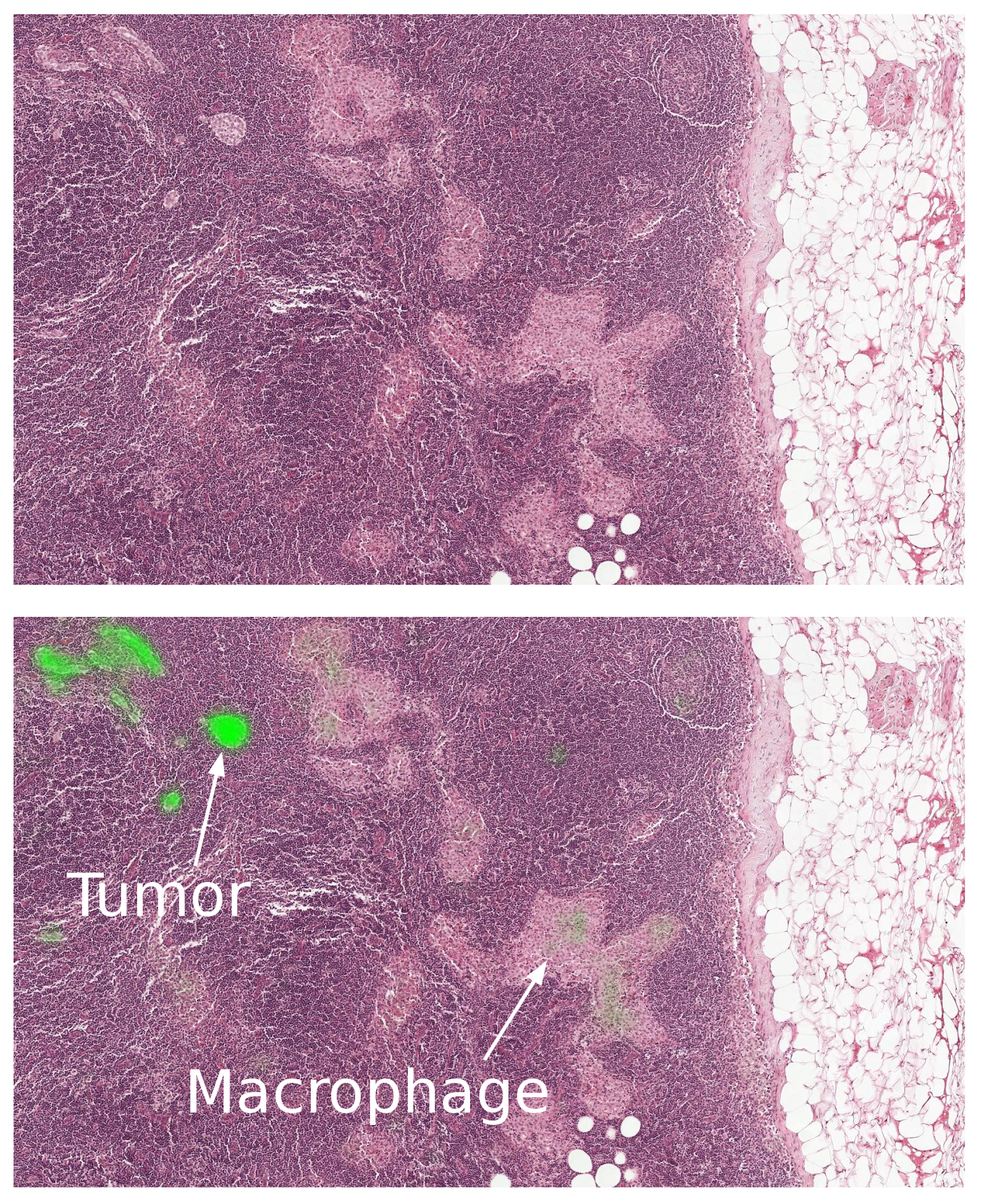 |
| A closeup of a lymph node biopsy. The tissue contains a breast cancer metastasis as well as macrophages, which look similar to tumor but are benign normal tissue. Our algorithm successfully identifies the tumor region (bright green) and is not confused by the macrophages. |
- Like most metrics, the FROC localization score is not perfect. Here, the FROC score is defined as the sensitivity (percentage of tumors detected) at a few pre-defined average false positives per slide. It is pretty rare for a pathologist to make a false positive call (mistaking normal cells as tumor). For example, the score of 73% mentioned above corresponds to a 73% sensitivity and zero false positives. By contrast, our algorithm’s sensitivity rises when more false positives are allowed. At 8 false positives per slide, our algorithms had a sensitivity of 92%.
- These algorithms perform well for the tasks for which they are trained, but lack the breadth of knowledge and experience of human pathologists — for example, being able to detect other abnormalities that the model has not been explicitly trained to classify (e.g. inflammatory process, autoimmune disease, or other types of cancer).
- To ensure the best clinical outcome for patients, these algorithms need to be incorporated in a way that complements the pathologist’s workflow. We envision that algorithm such as ours could improve the efficiency and consistency of pathologists. For example, pathologists could reduce their false negative rates (percentage of undetected tumors) by reviewing the top ranked predicted tumor regions including up to 8 false positive regions per slide. As another example, these algorithms could enable pathologists to easily and accurately measure tumor size, a factor that is associated with prognosis.
1 For those who might be interested, the Camelyon17 challenge, which builds upon the 2016 challenge, is currently underway.↩
2 The pathologist ended up spending 30 hours on this task on 130 slides.↩