
Overview
Digital pathology is changing the way pathology is practiced by making it easier to share images, collaborate with colleagues, and develop new AI algorithms that can improve the quality and cost of medical care. One of the biggest challenges of digital pathology is storing and managing the large volume of data generated. The Google Cloud Healthcare API provides a solution for this with a managed DICOM store, which is a secure, scalable, and performant way to store digital pathology images in a manner that is both standardized and interoperable.
However, performing image retrieval of specific patches (i.e. regions of interest) of a whole slide image (WSI) from the managed DICOM store using DICOMweb can be complex and requires DICOM format expertise. To address this, we are open sourcing EZ WSI (Whole Slide Image) DICOMWeb, a Python library that makes fetching these patches both efficient and easy-to-use.
How EZ WSI DICOMWeb works
EZ WSI DICOMweb facilitates the retrieval of arbitrary and sequential patches of a DICOM WSI from a DICOMWeb compliant Google Cloud Healthcare API DICOM store. Unlike downloading the entire DICOM series WSI and extracting patches locally from that file, which can increase network traffic, latency and storage space usage, EZ WSI DICOMweb retrieves only the necessary tiles for the desired patch directly through the DICOMweb APIs. This is simpler to use and abstracts away the following:
- The need to fetch many tiles, which requires an understanding of DICOM data structure (e.g. offset & data hierarchy).
- The need for a detailed understanding of the DICOMWeb APIs, REST payloads, and authentication, as well as addressing the possibility of redundant requests if several patches are fetched and there are overlapping tiles.
- The need to decode images on the server if client side decoding is not supported, which increases the time it takes to transfer data and the size of the data being transferred.
EZ WSI DICOMWeb allows researchers and developers to focus on their ML tasks rather than the intricacies of DICOM. Developers do not need to have an in-depth understanding of DICOM data structuring or the DICOM API. The library provides a simple and intuitive functionality that allows developers to efficiently fetch DICOM images using only the Google Cloud Platform (GCP) Resource Name and DICOM Series path without any pixel recompression.
Case Study: Generating Patches for AI Workflows
A typical pathology WSI could be on the order of 40,000 pixels in length or width. However, an AI model that is trained to assess that WSI may only analyze a patch that is 512 x 512 pixels at a time. The way the model can operate over the entire WSI is by using a sliding windows approach. We demonstrate how that can be done using EZ WSI DICOMWeb.
First, we create a DicomSlide object using the DICOMweb client and interface. This can be done with just a few lines of code.
dicom_web_client = dicom_web.DicomWebClientImpl()
dwi = dicom_web_interface.DicomWebInterface(dicom_web_client)
ds = dicom_slide.DicomSlide(
dwi=dwi,
path=gcp_resource_name+dicom_series_path,
enable_client_slide_frame_decompression = True
)
ds.get_image(desired_magnification) # e.g. '0.625X' |
This DicomSlide represents the entire WSI, as illustrated below.
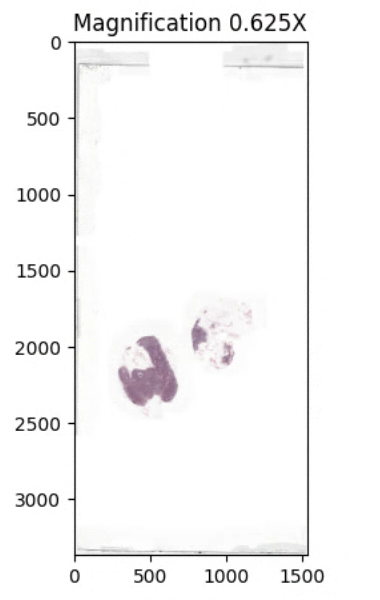 |
The above image leverages EZ WSI’s DicomSlide module to fetch an entire WSI at the requested magnification of 0.625X and uses matplotlib to render it, see the sample code for more details.
By providing coordinates, DicomSlide’s get_patch() method allows us to manually extract just the two sections of tissue at supported magnification with coordinates as pictured below.
tissue_patch = ds.get_patch(
desired_magnification, x=x_origin, y=y_origin, width=patch_width, height=patch_ height
) |
 |
We can effectively zoom in on patches programmatically by reducing the window size and increasing the magnification using the same get patch method from above.
 |
Our ultimate goal is to generate a set of patches that can be used in a downstream AI application from this WSI.
 |
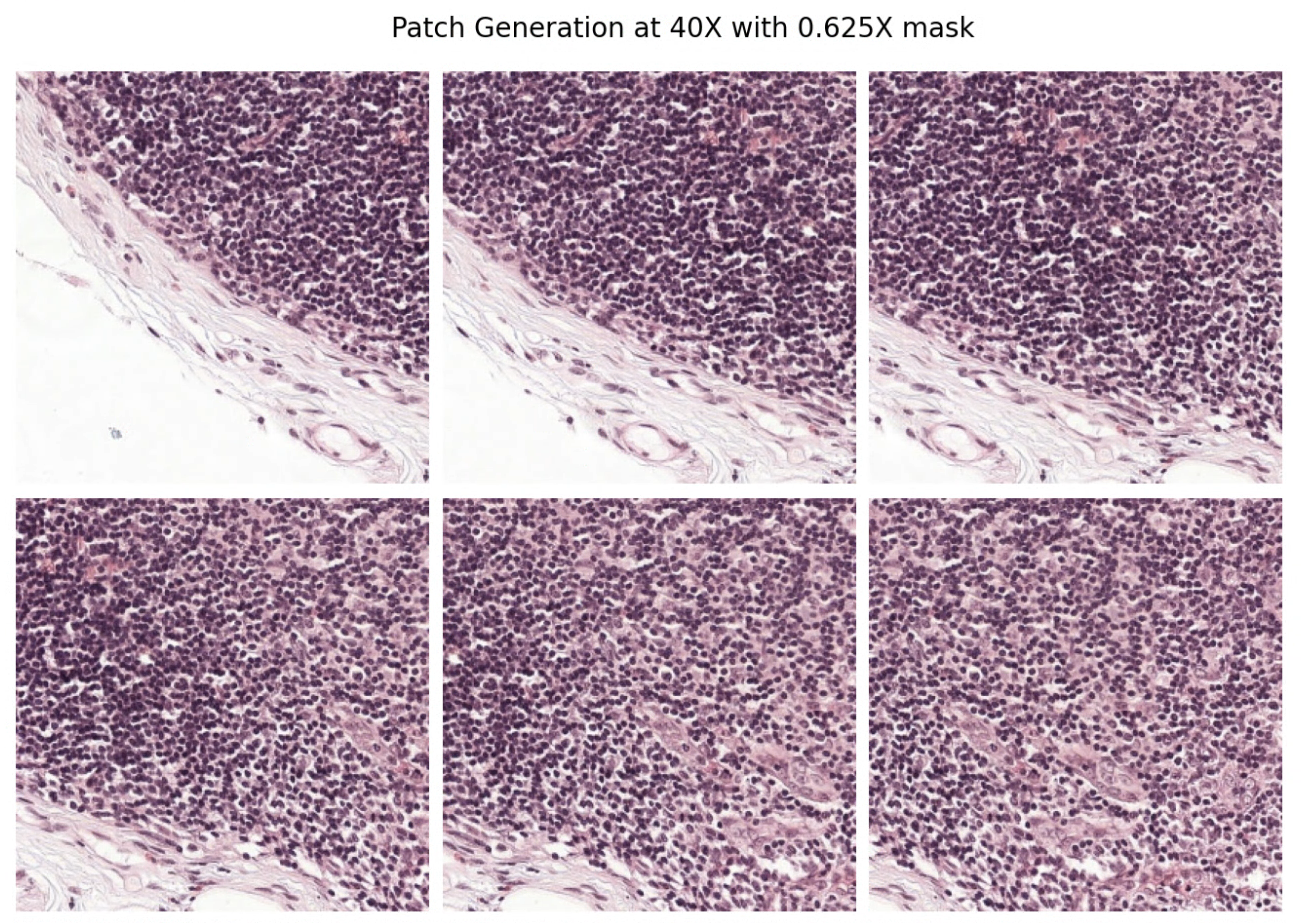
To do this, we call PatchGenerator. It works by sliding a window of a specified size with a specified stride size across the image, heuristically ignoring tissue-less regions at a specified magnification level.
patch_gen = patch_generator.PatchGenerator(
slide=ds,
stride_size=stride_size, # the number of pixels between patches
patch_size=patch_size, # the length and width of the patch in pixels
magnification=patch_magnification, # magnification to generate patches at
max_luminance=0.8, # defaults to .8, heuristic to evaluate where tissue is.
tissue_mask_magnification=mask_magnification,
) |
The result is a list of patches that can be used as input into a machine learning algorithm.
 |
Conclusion
We have built this library to make it easy to directly interact with DICOM WSIs that are stored in Google's DICOMWeb compliant Healthcare API DICOM store and extract image patches for AI workflows. Our hope is that by making this available, we can help accelerate the development of cutting edge AI for digital pathology in Google Cloud and beyond.
Links: Github, GCP-DICOMWeb
By Google HealthAI and Google Cloud Healthcare teams



